1 Introduction
Properly identifying the author of a scientific article is an important task for giving credit, tracking progress, and identifying ideas’ lineages. Usually, publications and citations do not provide unique identifiers to authors but only the raw string character representation of their name and affiliation. The fundamental problem is that an author might change the string representations due to changing in name spelling (e.g., removing accents), journal limitations (e.g., only allow first letter of first name), or simply two people having the same name. Several researchers have proposed methods to solve this problem[1,2,3], but most methods do not scale well and are not open to the community. In this work, we develop a scalable method that we make publicly available to disambiguate large-scale publications.
2 Methods
We proposed a method named ANNGC (Approximate Nearest Neighbors Graph Clustering), which contains three components: blocking, linkage, and clustering.
2.1 Blocking
The aim of blocking is to roughly partition S into a number of groups called blocks B = {b_1 b_2, ... b_k } and then perform disambiguation only within a block. By blocking, the computational complexity of a typical clustering algorithm O(|S|^2) is reduced to O(\sum{_{i=0}^{|B|}}|b_{i}|^{2}). By choosing |b_i|<<|S|, the difference in complexity is substantial.
2.2 Linkage
Training a function predicts the probability of two signatures belonging to the same author. In order to convert the signature pairs into feature vectors, we convert co-author names and abstract into vectors using word-wise tfidf, for the first name, full name, affiliation, title, and journal we use character-wise tfidf. We then compute the cosine similarity between those tfidf vectors of the signature pairs. We also use absolute year differences as features. All the features are fed into a logistic regression classifier.
2.3 Clustering
Several previous studies have used Hierarchical Agglomerative Clustering (HAC) to partition signatures into groups. . However, as the time complexity of HAC is O(n^3), it is difficult to scale to large blocks. Also, each merging step in HAC requires global distance information of the whole block, thus difficult to distribute the model into multiple nodes. In order to overcome these problems, we propose a scalable and distributable algorithm whose steps are described now (see Figure 1).
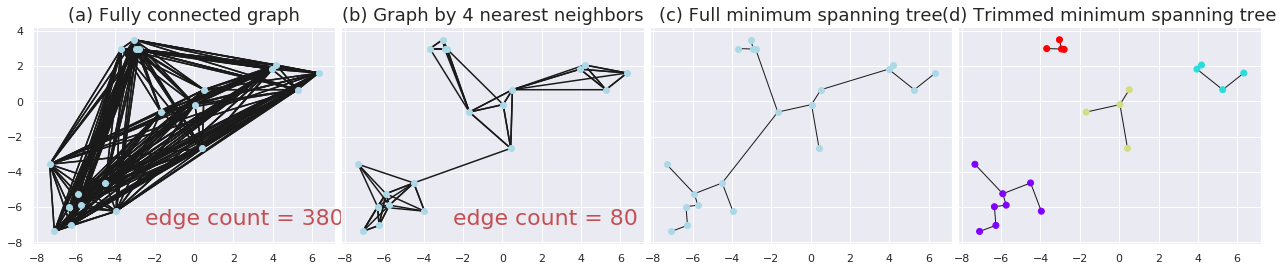
Please read the paper for explanation of each step.
2.4 Results
Baseline model: The HAC for clustering is used as a baseline. The blocking and linkage function are the same as ANNGC. As shown in Table 1, ANNGC local cut has the best precision, but the overall performance is slightly lower than the HAC. The local cut is better than global cut in both methods, as the thresholds are customized for each block.
| Description | Precision | Recall | F1 |
|---|---|---|---|
| Baseline-Global cut | 0.978 | 0.9793 | 0.9788 |
| Baseline-local cut | 0.997 | 0.9946 | 0.9962 |
| ANNGC-Global cut | 0.968 | 0.9766 | 0.9727 |
| ANNGC-local cut | 0.998 | 0.9523 | 0.9747 |
2.5 Conclusion
While the performance of ANNGC is slightly lower than HAC, our approach has three advantages: 1) less time complexity. The complexity is O(V·h·h_root(V)) for ANN plus O(E·logV) for MST, much lower than HAC (O(V^3)), given V the number of vertices, E the number of edges. 2) distributable, all the steps (ANN, MST, connected component) could be implemented without having the information of the whole blocks. 3) fewer hyper-parameters. There are only two parameters, the k for nearest neighbors and the cut-off threshold. These features are critical for large scale author name disambiguation which is not capable for HAC. In the future, we will experiment with network embeddings which could reduce our vector dimension from million to thousands. Also, we could use more information about the paper, such as the reference list. We could also apply more advanced models for the linkage function.
Citation
@inproceedings{zeng2020,
author = {Zeng, Tong and E Acuna, Daniel},
title = {Large-Scale Author Name Disambiguation Using Approximate
Network Structures},
booktitle = {6th International Conference on Computational Social
Science},
date = {2020-07-17},
eventdate = {},
url = {https://tong-zeng.github.io/publications/large-scale-author-name-disambiguation/},
langid = {en},
abstract = {Properly identifying the author of a scientific article is
an important task for giving credit, tracking progress, and
identifying ideas’ lineages. Usually, publications and citations do
not provide unique identifiers to authors but only the raw string
character representation of their name and affiliation. The
fundamental problem is that an author might change the string
representations due to changing in name spelling (e.g., removing
accents), journal limitations (e.g., only allow first letter of
first name), or simply two people having the same name. Several
researchers have proposed methods to solve this problem, but most
methods do not scale well and are not open to the community. In this
work, we develop a scalable method that we make publicly available
to disambiguate large-scale publications.}
}